EVOLUTIONARY FOOTPRINTS OF THE REPLICATION(S) MECHANISM(S) IN VERTEBRATE mtDNA
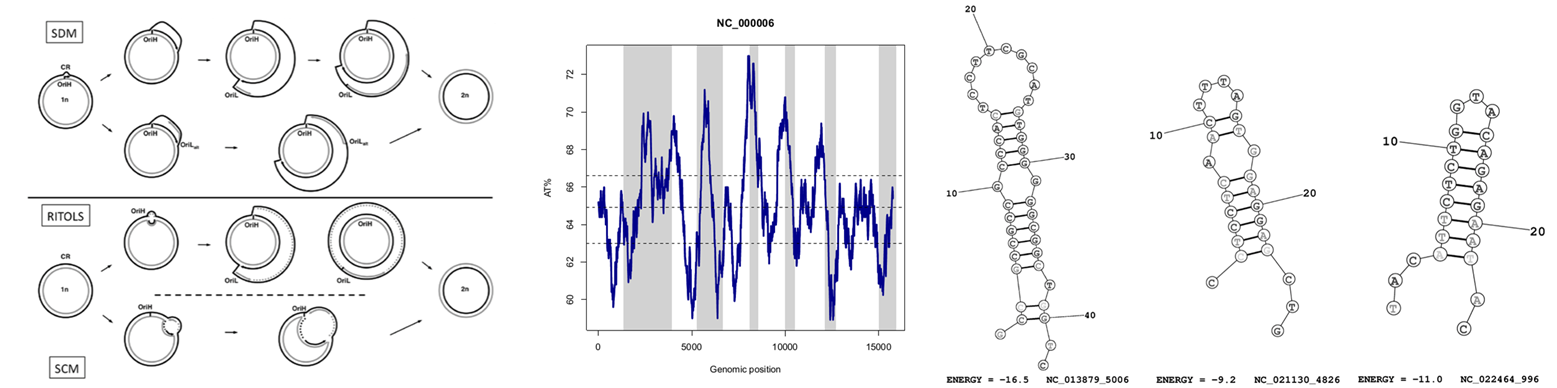
Mitochondrial genomes are known to have a strong strand-specific compositional bias that is more pronounced at fourfold redundant sites of mtDNA protein-coding genes. This observation suggests that strand asymmetries, to a large extent, are caused by mutational asymmetric mechanisms. In vertebrate mitogenomes, replication and not transcription seems to play a major role in shaping compositional bias. Hence, one can better understand how mtDNA is replicated through a detailed picture of mitochondrial genome evolution. Although at the biochemical level the model for vertebrate mtDNA replication is still a debated issue, from a molecular evolutionary perspective, several features of vertebrate mtDNA are traditionally associated with a strand-displacement model. In this seminar I will present our latest findings on the role of replication in mtDNA evolution based on the analysis of almost 2,500 complete mitochondrial genomes.
Miguel Fonseca graduated in Biology at the University of Aveiro (Portugal) in 2004. He then completed a MSc degree in Biodiversity and Genetic Resources (CIBIO-InBIO/University of Porto, Portugal). During this time, his work was mainly focused on the phylogenetic and phylogeographic history of North African lizards of the genus Acanthodactylus. Simultaneously, he started doing some research related to the general evolution of mitochondrial DNA. Specifically, he was interested in testing if mitochondrial gene rearrangements could leave an evolutionary signal at the DNA level. During his PhD (completed in 2011, University of Porto), he carried out molecular and comparative genomics analyses on the evolution of vertebrate mitochondrial DNA. Presently, Miguel is involved in different projects (phylogenomics and comparative genomics) that use mitogenomes as an informative molecule and he is also applying molecular (time and spatial) heterogeneous models to correct systematic errors that occur in phylogenetics inference.
[Group Leader: Nuno Ferrand de Almeida, Population Genetics, Hybridization and Speciation]
Image credits: Miguel Fonseca