PREY DNA DETECTABILITY HALF-LIVES: TURNING PCR POSITIVES INTO QUANTITATIVE PREDATION RATES
Predation affects every facet of species ecology, either in the context of being a predator or being prey, including distribution, resource partitioning, population fluxes, circadian rhythms and evolution. Furthermore, predation influences important conservation and resource management issues such as the impact of introduced fauna on native prey or the impact of pest predators on commercially valued prey (e.g. pollinators, game or fish).
Diet analysis remains one of the most important tools in predation studies and DNA-based methods have largely surpassed traditional morphological diet analysis as a means of detecting prey items in animal gastrointestinal and faecal samples. Usually, DNA-based diet analysis results in qualitative (or semi-quantitative) Frequency of Occurrence data. Depending on the objective, these data alone can be useful, but where the objective is to measure the impact of one species on another, quantitative predation rates are necessary.
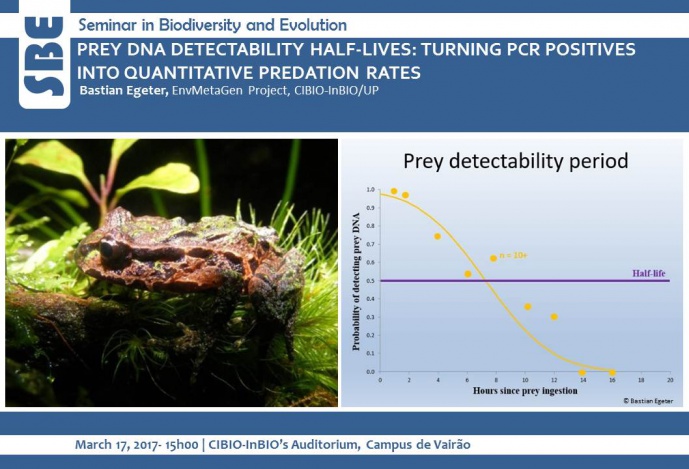
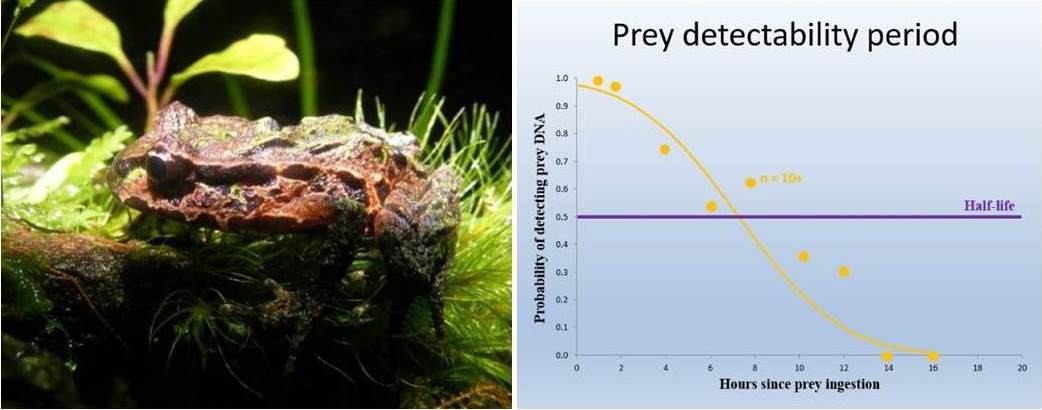
Here I present a case study that combined DNA-based diet analysis, prey detectability half-lives, predator density and prey availability to estimate the impact of introduced ship rats (Rattus rattus) on New Zealand´s Archey´s frog (Leiopelma archeyi), which is currently ranked as the world´s most endangered amphibian (ZSL Amphibian EDGE list). Estimates of predation ranged from 0.07-0.3 frogs consumed per rat per night, information that was utilised by the Department of Conservation in the decision making process on providing rodent control. The study represents the first to measure prey DNA detectability half-lives in mammal stomachs, the first to compare half-lives of stomach and faecal contents for any vertebrate, and the first to report a quantitative predation rate from standard PCR diet analysis.
Bastian joined CIBIO-InBIO last December as a post-doc in the EnvMetaGen project to work on a range of molecular ecology studies. He is currently involved in an amphibian metabarcoding project that will investigate the diversity of amphibians using eDNA obtained from Mauritanian gueltas (closely linked with the BIODESERT research group). He has also recently started a project investigating the prevalence and diversity of Phosphorus-freeing genes in soil, with the overarching aim of improving the productivity of agricultural systems.
Prior to coming here Bastian´s research has always had a molecular ecology focus, with topics including frog conservation, invasive mammal ecology, plant pathogen detection and Antarctic nematode ecology. On the conservation frontline he has worked as an ecological consultant focussing on badgers, otters, amphibians and lizards.
[Host: Simon Jarman, EnvMetaGen Project]
Image credits: Bastian Egeter