ESTIMATING INBREEDING FROM LOW-COVERAGE NGS DATA
WELCOME SEMINAR IN BIODIVERSITY AND EVOLUTION
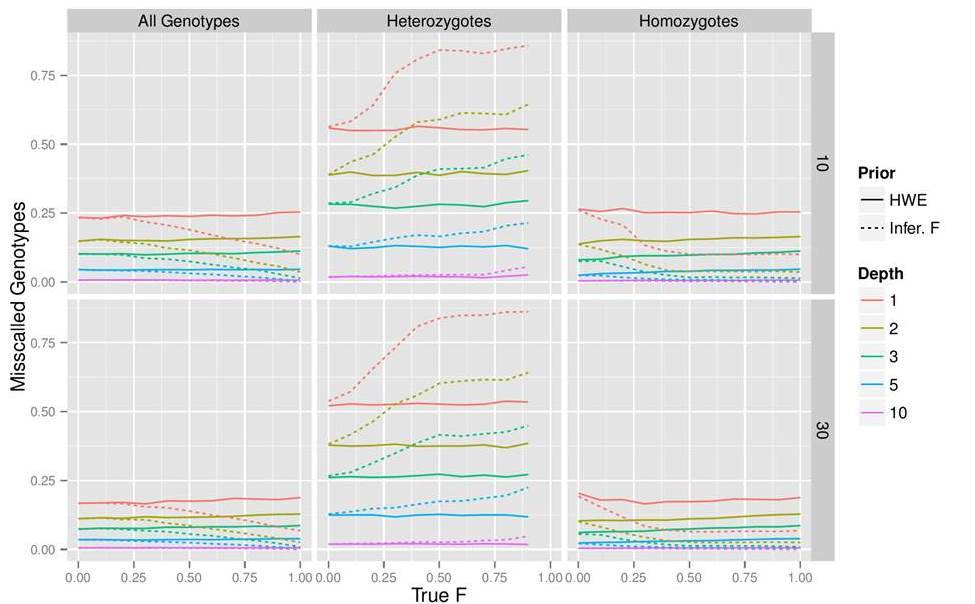
Current NGS technologies produce short read sequences that are de-novo assembled or mapped (aligned) to a reference genome and used for SNP or genotype calling. However, these data typically have high error rates due to multiple factors, from random sampling of homologous base pairs in heterozygotes, to sequencing or alignment errors. Furthermore, many NGS studies rely on low coverage sequence data (< 5× per site per individual), causing SNP and genotype calling to be associated with considerable statistical uncertainty.
Recent methods rely on probabilistic frameworks to account for these errors and accurately call SNPs and genotypes, even at low depths. These methods integrate the base quality score together with other error sources (e.g., mapping or sequencing errors) to calculate an overall ”genotype likelihood”. This likelihood function can be combined with a prior to calculate a posterior probability for the genotype. Most genotype calling methods for Next Generation Sequencing (NGS) data use priors based on allele frequencies under the assumption of Hardy-Weinberg Equilibrium (HWE). However, many organisms including domesticated, partially selfing or with asexual life cycles show strong HWE deviations. For such species, and specially with low coverage data, it is necessary to obtain estimates of inbreeding coefficients for each individual before any further analyses.
Coming from the University of California Berkeley, Filipe Vieira has recently joined CIBIO-InBIO to work as part of the new bioinformatics group. Filipe holds a PhD in comparative genomics, from the University of Barcelona. During his doctoral studies, Filipe focused on the study of the evolution of multigenic families in insects. Afterwards, he worked as a Postdoc on population genetics/genomics, with a special emphasis on Next-Generation Sequencing (NGS) data. In this scope, Filipe worked mainly on two topics: i) the development of new NGS methods, which will be main focus of this introductory talk; and ii) the study of rice domestication.
Image credits: Filipe Vieira