ADVANCED COURSE: MEASURING AND MAPPING EVOLUTIONARY DIVERSITY FOR CONSERVATION PLANNING
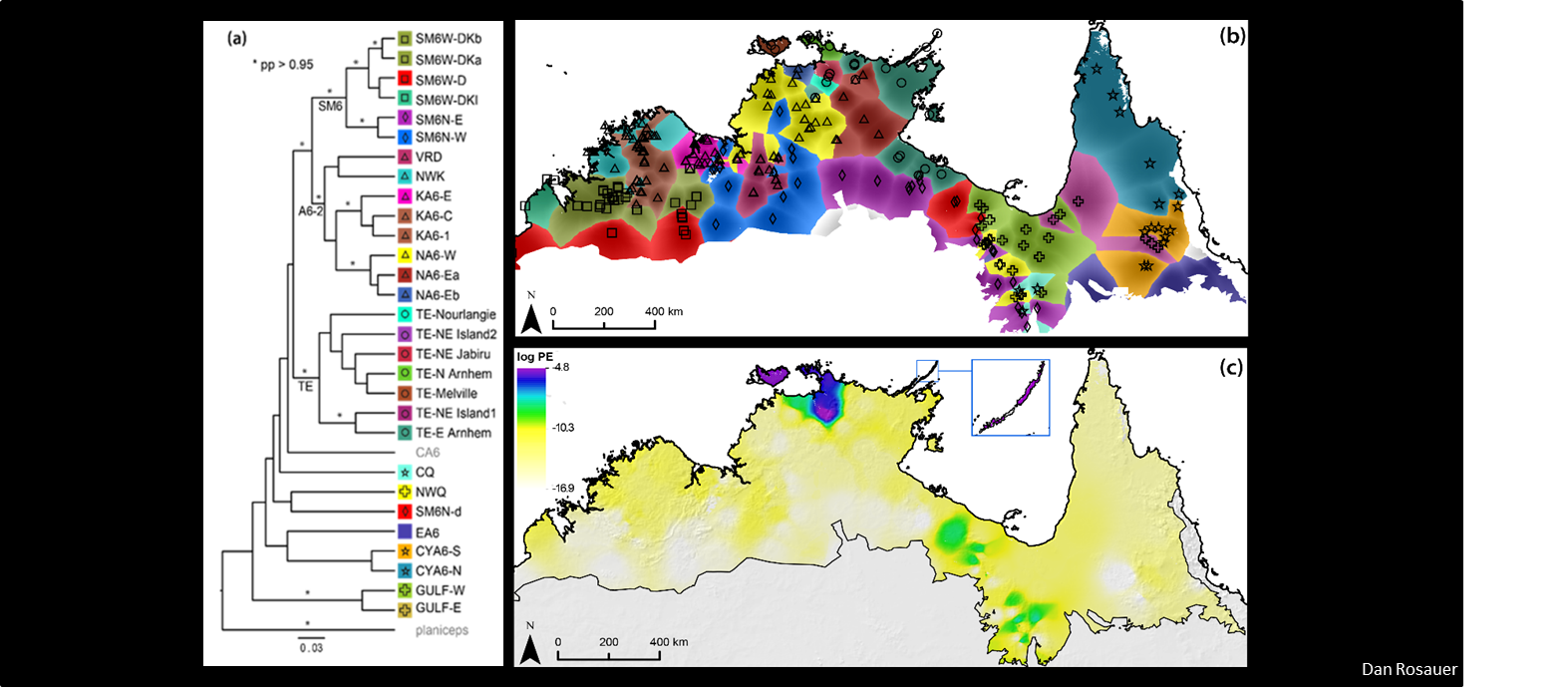
There is a growing recognition of the importance of accounting for evolutionary relationships when prioritizing conservation actions, however in practice, they are rarely considered. Using phylogenies we can calculate a set of metrics to quantify evolutionary diversity within or among a set of taxa. Recently, several methods and tools have emerged to apply phylogenetic information to measure and map biodiversity for conservation.
At the end of this course, participants will be able to:
- Identify different facets and dimensions of biodiversity;
- Understand how biodiversity conservation can benefit from accounting for evolutionary relationships within and between species;
- Recognize the multiple available metrics to measure evolutionary information and how they relate to each other;
- Understand which metrics to use for particular conservation problems;
- Perform essential data manipulation in R using spatial data and phylogenetic trees;
- Calculate and map phylogenetic diversity, phylogenetic endemism and evolutionary distinctiveness;
- Understand the importance of null models in detecting spatial patterns of phylogenetic diversity; and
- Predict the distribution of sub-specific phylogeographic lineages using interpolation and modelling approaches.
PROGRAMME
May 8th
9h45 – 12h45
Introduction (Theory)
- What is biodiversity and why is it important
- Phylogenies - what they mean, how are they inferred, how to read them
- Measures and dimensions of biodiversity
- Why preserve evolutionary history and genetic diversity?
- Quantifying evolutionary history
- Richness, rarity and regularity metrics
14h45-17h45
Phylogenetic data in R (Hands on)
- Sources of phylogenetic trees
- Loading phylogenies
- Plotting phylogenetic trees
- Manipulating and calculating phylogenetic metrics from phylogenetic trees
Spatial Data in R (Hands on)
- Types of spatial data in R
- Loading and manipulating spatial data in R
- Basic spatial operations
May 9th
9h45 – 12h45
Using phylogenetic trees in conservation (Theory)
- Why is phylogenetics important for conservation?
- Prioritizing species based on evolutionary distinctiveness – the EDGE program
- Prioritizing sites based on phylogenetic metrics: mapping phylogenetic diversity and phylogenetic endemism
An introduction to null models for phylogenetic analysis (Theory)
- Why use null models for phylogenetic analysis
- Calculating standardized effect sizes, quantiles, and p-values
14h45-17h45
Mapping evolutionary diversity (Hands on)
- Phylogenetic diversity
- Evolutionary distinctiveness
- Phylogenetic endemism
- Using Biodiverse to visualize spatial and phylogenetic patterns
Analyzing phylogenetic data using null models (Hands on)
May 10th
9h45 – 12h45
Diversity below the species level and its importance for conservation (Theory)
Mapping phylogeographic lineages using an interpolation approach (Theory)
- General view on interpolation approaches with genetic distance
- Use of variograms to relate genetic distance with space
- Fitting models to variograms and interpolation
- Accounting for geography, landscape and environment in the interpolation
14h45-17h45
Mapping phylogeographic lineages with phylin (Hands on)
May 11th
9h45 – 12h45
A modelling approach to map phylogeographic lineages (Hands on)
- Modelling phylogeography lineages using habitat connectivity
- Mapping endemism from lineage models
14h45-17h45
Discussion and Perspectives
COURSE INSTRUCTORS
Silvia Carvalho - CIBIO-InBIO | THEOECO
Daniel Rosauer - Australian National University
Pedro Tarroso - CIBIO - InBIO | BIODESERTS
INTENDED AUDIENCE
The course will be open to a maximum number of 25 participants.
Priority will be given to:
- 1st year and other PhD students attending the BIODIV Doctoral Program;
- PhD students attending other courses;
- Other post-graduate students and researchers.
REQUIREMENTS
The software used for this course will be centered on the R language for statistics R: www.r-project.org. Basic knowledge of R and statistics is expected.
Participants should bring their personal laptop with the following software installed: R and packages ape, picante, raster and phylin
REGISTRATION
Registration deadline: April 1, 2017.
Participation is free of charge for BIODIV students | 80 € (students) / 150 € (other participants). CIBIO members will have an additional discount of 20%. Does not include lunch nor coffee breaks.
To register, please send an e-mail accompanied by your short CV (max. two A4 pages) to post.graduation@cibio.up.pt. Please refer your status (PhD student, MSc Student, Other) and the University to which you are affiliated.
![]()